Chapter 1 Data handling with R
1.1 Data types
- numeric:
212.125
- character strings:
'a',"word" - logical:
TRUEFALSENULL(sort of)
Please note that logicals are in capital letters.
- vectors:
c(12, 15, 35698)c("FOSS4G 2022", "Firenze")
- list:
list(1, 45, 12.0, "toto") - matrices:
matrix(0:9, 3,3) - data frames (df):
data.frame(x = 1:3, y = c("a", "b", "c")) - constants:
letters[3:4]LETTERS[12:26]pi
1.3 Not only a calculator
R is shipped with lots of functions like :
data(<datasetname>) # load an embedded dataset
head(<objectname>) # first lines of a dataframe
is.vector(object) # return TRUE if object is a vector
is.data.frame(object) # return TRUE if object is a data.frame
class(<objectname>) # returns the class of an object
unique(<objectname>) # returns unique values
help(<functionname>) # get help on a function
plot(<objectname>) # create a graphic from a datasetExercise:
- Load the
LakeHurondataset usingdata() - Get help on the
LakeHurondataset - Use
head()to see its first lines - Plot
LakeHuron
1.4 Packages
While base R contains a lot of functions, it can be extended with various packages.
In R, the fundamental unit of shareable code is the package. A package bundles together code, data, documentation, and tests, and is easy to share with others. Wickham and Bryan (2022)
You already saw how to install packages with the install.packages() function, let’s see how to load the {dplyr}
package we will use to handle the data.
1.5 Load data
If the packages do not provide datasets to work with, you will want to work on your own data. There is several ways to load data.
Base R provides a set of functions for delimited text files. For example, if we want to work with the Gapminder dataset from Software Carpentry’s R course.
data_url <- "https://raw.githubusercontent.com/swcarpentry/r-novice-gapminder/main/data/gapminder_data.csv"
# download the data and store it in a variable
gapminder <- read.csv(data_url)We call this dataset from an URL but it can be a path in your system file.
For example, if you have installed the {gapminder} package as recommended, you will find the data set as a TSV file in your R installation.
## [1] "/Users/runner/work/_temp/Library/gapminder/extdata/gapminder.tsv"system.file() is a function that returns the path of files in R packages, independently
of the operating system.
read.csv() and similar functions can read delimited text files only.
For other formats, you can use functions from other packages like {haven} or {readxl}.
We will show in Chapter 2 how to load geospatial data.
Let’s take a look to our data :
## Rows: 1,704
## Columns: 6
## $ country <chr> "Afghanistan", "Afghanistan", "Afghanistan", "Afghanistan", …
## $ year <int> 1952, 1957, 1962, 1967, 1972, 1977, 1982, 1987, 1992, 1997, …
## $ pop <dbl> 8425333, 9240934, 10267083, 11537966, 13079460, 14880372, 12…
## $ continent <chr> "Asia", "Asia", "Asia", "Asia", "Asia", "Asia", "Asia", "Asi…
## $ lifeExp <dbl> 28.801, 30.332, 31.997, 34.020, 36.088, 38.438, 39.854, 40.8…
## $ gdpPercap <dbl> 779.4453, 820.8530, 853.1007, 836.1971, 739.9811, 786.1134, …This dataset is about life expectancy, population and GDP per capita in world countries between 1952 and 2007.
You can also use head() for the same purpose but a different output format.
## country year pop continent lifeExp gdpPercap
## 1 Afghanistan 1952 8425333 Asia 28.801 779.4453
## 2 Afghanistan 1957 9240934 Asia 30.332 820.8530
## 3 Afghanistan 1962 10267083 Asia 31.997 853.1007
## 4 Afghanistan 1967 11537966 Asia 34.020 836.1971
## 5 Afghanistan 1972 13079460 Asia 36.088 739.9811
## 6 Afghanistan 1977 14880372 Asia 38.438 786.1134We can transform a data frame into a tibble to access their better print() method that combines head() and glimpse().
Tibbles are at the core of the Tidyverse packages.
## # A tibble: 1,704 × 6
## country year pop continent lifeExp gdpPercap
## <chr> <int> <dbl> <chr> <dbl> <dbl>
## 1 Afghanistan 1952 8425333 Asia 28.8 779.
## 2 Afghanistan 1957 9240934 Asia 30.3 821.
## 3 Afghanistan 1962 10267083 Asia 32.0 853.
## 4 Afghanistan 1967 11537966 Asia 34.0 836.
## 5 Afghanistan 1972 13079460 Asia 36.1 740.
## 6 Afghanistan 1977 14880372 Asia 38.4 786.
## 7 Afghanistan 1982 12881816 Asia 39.9 978.
## 8 Afghanistan 1987 13867957 Asia 40.8 852.
## 9 Afghanistan 1992 16317921 Asia 41.7 649.
## 10 Afghanistan 1997 22227415 Asia 41.8 635.
## # … with 1,694 more rowsIf you want more information about the gapminder dataset, Hans Rosling made a TED talk presenting the gapminder data.
We can see that it contains several columns (also called variables):
## [1] "country" "year" "pop" "continent" "lifeExp" "gdpPercap"1.6 In the beginning was the Verb
{dplyr} provides a lot of functions to handle data: filter() to filter the data matching certain conditions (subset data), select() to select columns, mutate() to create new variables.
All those functions are verbs and means an action onto the dataset.
1.7 Filter data
If we only want the records from Italy, we can filter using:
## # A tibble: 12 × 6
## country year pop continent lifeExp gdpPercap
## <chr> <int> <dbl> <chr> <dbl> <dbl>
## 1 Italy 1952 47666000 Europe 65.9 4931.
## 2 Italy 1957 49182000 Europe 67.8 6249.
## 3 Italy 1962 50843200 Europe 69.2 8244.
## 4 Italy 1967 52667100 Europe 71.1 10022.
## 5 Italy 1972 54365564 Europe 72.2 12269.
## 6 Italy 1977 56059245 Europe 73.5 14256.
## 7 Italy 1982 56535636 Europe 75.0 16537.
## 8 Italy 1987 56729703 Europe 76.4 19207.
## 9 Italy 1992 56840847 Europe 77.4 22014.
## 10 Italy 1997 57479469 Europe 78.8 24675.
## 11 Italy 2002 57926999 Europe 80.2 27968.
## 12 Italy 2007 58147733 Europe 80.5 28570.in Tidyverse functions, the data is the first argument and does not need to be named.
You can use comparison operators like ==, >, <, >= , etc.
There is also logical operators : & (AND), | (OR), ! (NOT) and xor().
So, for example, if we want to subset records for Italy after 2000, we can use filter like this:
## # A tibble: 2 × 6
## country year pop continent lifeExp gdpPercap
## <chr> <int> <dbl> <chr> <dbl> <dbl>
## 1 Italy 2002 57926999 Europe 80.2 27968.
## 2 Italy 2007 58147733 Europe 80.5 28570.This can be translated to : “From the gapminder dataset, filter all rows where the country is equal to Italy and the year is superior to 2000”.
1.8 Select columns
select() allows you to keep only the columns you need for your analysis.
Maybe we only want the country, year and lifeExp variables:
## # A tibble: 2 × 3
## country year lifeExp
## <chr> <int> <dbl>
## 1 Italy 2002 80.2
## 2 Italy 2007 80.5Or, for example, in the italy_2000 subset, the continent variable does not provide useful information anymore, so we want to discard it.
Please not the use of the - symbol before the name of the variable.
## # A tibble: 2 × 5
## country year pop lifeExp gdpPercap
## <chr> <int> <dbl> <dbl> <dbl>
## 1 Italy 2002 57926999 80.2 27968.
## 2 Italy 2007 58147733 80.5 28570.So you can select the column you want to keep or the ones you want to remove.
1.9 Create new variables
Let say you want to compute the GDP, in millions, from the population and the GDP per capita variables.
For that, you can use the mutate() function:
## # A tibble: 1,704 × 7
## country year pop continent lifeExp gdpPercap GDP
## <chr> <int> <dbl> <chr> <dbl> <dbl> <dbl>
## 1 Afghanistan 1952 8425333 Asia 28.8 779. 6567.
## 2 Afghanistan 1957 9240934 Asia 30.3 821. 7585.
## 3 Afghanistan 1962 10267083 Asia 32.0 853. 8759.
## 4 Afghanistan 1967 11537966 Asia 34.0 836. 9648.
## 5 Afghanistan 1972 13079460 Asia 36.1 740. 9679.
## 6 Afghanistan 1977 14880372 Asia 38.4 786. 11698.
## 7 Afghanistan 1982 12881816 Asia 39.9 978. 12599.
## 8 Afghanistan 1987 13867957 Asia 40.8 852. 11821.
## 9 Afghanistan 1992 16317921 Asia 41.7 649. 10596.
## 10 Afghanistan 1997 22227415 Asia 41.8 635. 14122.
## # … with 1,694 more rowsIt’s companion function, transmute() does the same thing but only keep the new variables.
## # A tibble: 1,704 × 1
## GDP
## <dbl>
## 1 6567.
## 2 7585.
## 3 8759.
## 4 9648.
## 5 9679.
## 6 11698.
## 7 12599.
## 8 11821.
## 9 10596.
## 10 14122.
## # … with 1,694 more rowsIf you want to keep variables from the dataset, you can call them in the function call:
## # A tibble: 1,704 × 3
## country year GDP
## <chr> <int> <dbl>
## 1 Afghanistan 1952 6567.
## 2 Afghanistan 1957 7585.
## 3 Afghanistan 1962 8759.
## 4 Afghanistan 1967 9648.
## 5 Afghanistan 1972 9679.
## 6 Afghanistan 1977 11698.
## 7 Afghanistan 1982 12599.
## 8 Afghanistan 1987 11821.
## 9 Afghanistan 1992 10596.
## 10 Afghanistan 1997 14122.
## # … with 1,694 more rowsSo in this example, we compute the GDP but we also keep the information about the country and the year.
1.10 Agregate data
Sometimes, we have a too detailed dataset and we want a more broader view of the data so we want to aggregate it.
To do so, {dplyr} provides a couple of functions: group_by() and summarise().
Like their names say, group_by() groups records which share the same value in a variable and summarise() compute the summary of non-grouping variables.
For example, let’s compute the yearly population of each continent.
To do this, we first group the data by continent and year and pass the result to summarise().
In this second function, we create a new variable called population that is the sum of the variable pop of each group.
## `summarise()` has grouped output by 'continent'. You can override using the
## `.groups` argument.## # A tibble: 60 × 3
## # Groups: continent [5]
## continent year population
## <chr> <int> <dbl>
## 1 Africa 1952 237640501
## 2 Africa 1957 264837738
## 3 Africa 1962 296516865
## 4 Africa 1967 335289489
## 5 Africa 1972 379879541
## 6 Africa 1977 433061021
## 7 Africa 1982 499348587
## 8 Africa 1987 574834110
## 9 Africa 1992 659081517
## 10 Africa 1997 743832984
## # … with 50 more rowsYou can use any function to summarise your data, for example, if you want to know the number of entries by continent, you can use n().
## # A tibble: 5 × 2
## continent count
## <chr> <int>
## 1 Africa 624
## 2 Americas 300
## 3 Asia 396
## 4 Europe 360
## 5 Oceania 24Or if you only want the first country of each continent:
## # A tibble: 5 × 2
## continent country
## <chr> <chr>
## 1 Africa Algeria
## 2 Americas Argentina
## 3 Asia Afghanistan
## 4 Europe Albania
## 5 Oceania Australia.data is a pronoun that you can use when the column name is a character vector.
1.11 Join data
{dplyr} provides a large variety of functions to join datasets :
inner_join(),left_join(), right_join(), full_join(), nest_join() ,
semi_join(),anti_join().
Let’s create two datasets that shares a common key. This new key will be the country name and the year separated by an underscore.
gapminder_left <- transmute(gapminder,
key = paste0(country, "_" , year),
country,
continent,
year)
gapminder_left## # A tibble: 1,704 × 4
## key country continent year
## <chr> <chr> <chr> <int>
## 1 Afghanistan_1952 Afghanistan Asia 1952
## 2 Afghanistan_1957 Afghanistan Asia 1957
## 3 Afghanistan_1962 Afghanistan Asia 1962
## 4 Afghanistan_1967 Afghanistan Asia 1967
## 5 Afghanistan_1972 Afghanistan Asia 1972
## 6 Afghanistan_1977 Afghanistan Asia 1977
## 7 Afghanistan_1982 Afghanistan Asia 1982
## 8 Afghanistan_1987 Afghanistan Asia 1987
## 9 Afghanistan_1992 Afghanistan Asia 1992
## 10 Afghanistan_1997 Afghanistan Asia 1997
## # … with 1,694 more rowsgapminder_right <- transmute(gapminder,
key = paste0(country, "_" , year),
lifeExp,
pop,
gdpPercap)
gapminder_right## # A tibble: 1,704 × 4
## key lifeExp pop gdpPercap
## <chr> <dbl> <dbl> <dbl>
## 1 Afghanistan_1952 28.8 8425333 779.
## 2 Afghanistan_1957 30.3 9240934 821.
## 3 Afghanistan_1962 32.0 10267083 853.
## 4 Afghanistan_1967 34.0 11537966 836.
## 5 Afghanistan_1972 36.1 13079460 740.
## 6 Afghanistan_1977 38.4 14880372 786.
## 7 Afghanistan_1982 39.9 12881816 978.
## 8 Afghanistan_1987 40.8 13867957 852.
## 9 Afghanistan_1992 41.7 16317921 649.
## 10 Afghanistan_1997 41.8 22227415 635.
## # … with 1,694 more rowsNow we can join them.
joined_gapminder <- left_join(
gapminder_left,
gapminder_right,
by = "key" # optional argument if the join variables have the same name
)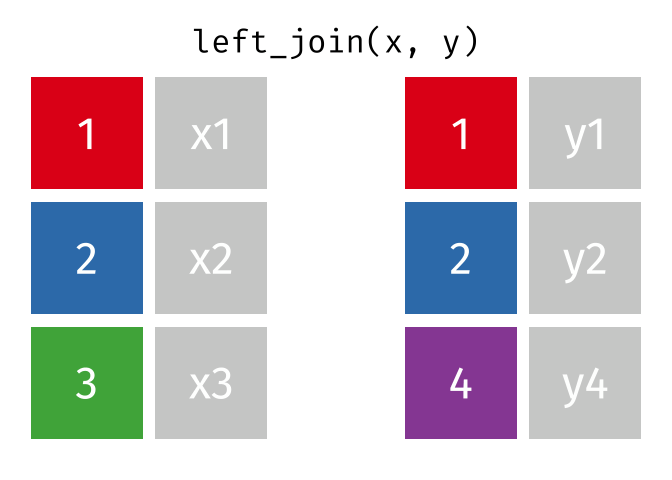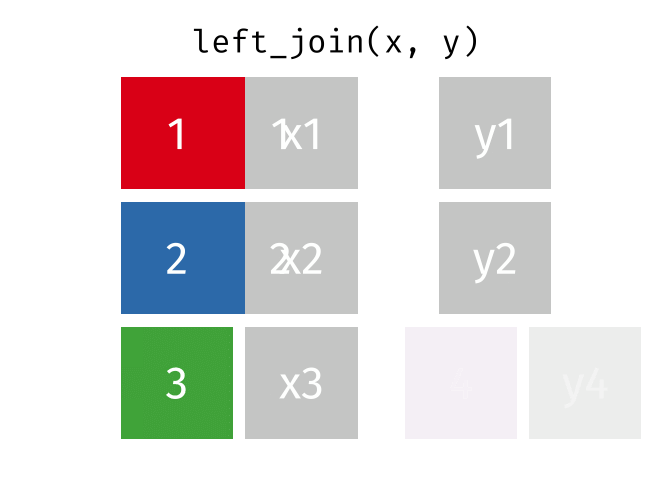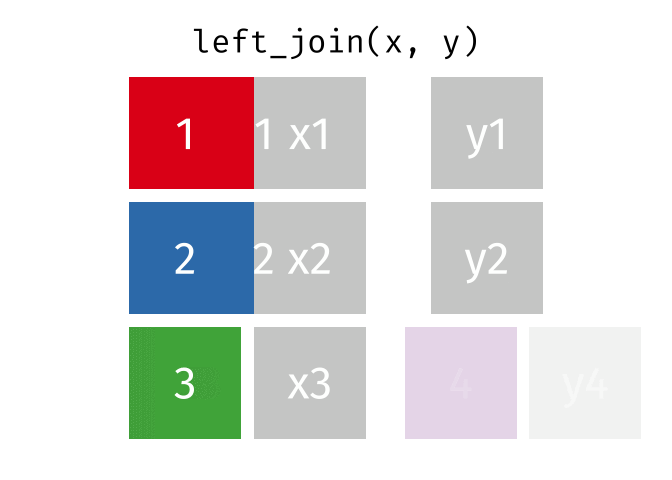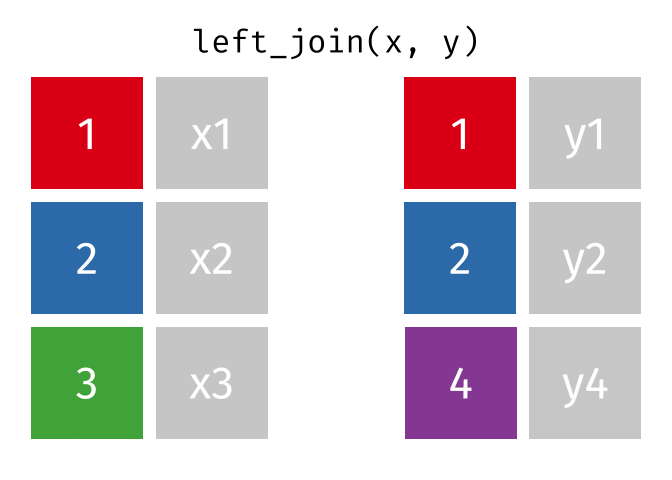
Left Join animation (Copyright Garrick Aden_Buie)
If you want more information on joins operations with {dplyr}, we recommend to read the dedicated blogpost from Garrick Aden-Buie
1.12 Piping
Piping allows to create a sequence of actions on a dataset without storing intermediate results.
As it can be difficult to debug piped commands for beginners, we won’t use it in this workshop. However, its usage is very frequent so it is most likely that a beginner will encounter it in documentation or in code source publicly available.
The most common form %>% is provided by the package {magrittr}, which is part of the tidyverse and is a dependency of {dplyr}, so you don’t have to load it.
In the following example, we show how to compute the mean GDP by the decade of European countries using pipes to chain functions.
gapminder %>% # pass the dataset as the first argument
filter(continent == "Europe") %>% # subset on European records
select(-continent) %>% # remove the continent column
mutate(decade = year - year %% 10, # compute decade
GDP = (gdpPercap * pop) / 1000000 # compute GDP
) %>%
group_by(country, decade) %>% # grouping variables
summarise(mean_GDP = mean(GDP)) # compute mean GDP of the decade## `summarise()` has grouped output by 'country'. You can override using the
## `.groups` argument.## # A tibble: 180 × 3
## # Groups: country [30]
## country decade mean_GDP
## <chr> <dbl> <dbl>
## 1 Albania 1950 2461.
## 2 Albania 1960 4737.
## 3 Albania 1970 8182.
## 4 Albania 1980 10796.
## 5 Albania 1990 9627.
## 6 Albania 2000 18765.
## 7 Austria 1950 52056.
## 8 Austria 1960 85666.
## 9 Austria 1970 137585.
## 10 Austria 1980 171559.
## # … with 170 more rowsWith R 4.1.0, the built-in piping operator |> has been introduced:
gapminder |> # pass the datset as the first argument
filter(continent == "Europe") |> # subset on European records
select(-continent) |> # remove the continent column
mutate(decade = year - year %% 10, # compute decade
GDP = (gdpPercap * pop) / 1000000 # compute GDP
) |>
group_by(country, decade) |> # grouping variables
summarise(mean_GDP = mean(GDP)) # compute mean GDP of the decade## `summarise()` has grouped output by 'country'. You can override using the
## `.groups` argument.## # A tibble: 180 × 3
## # Groups: country [30]
## country decade mean_GDP
## <chr> <dbl> <dbl>
## 1 Albania 1950 2461.
## 2 Albania 1960 4737.
## 3 Albania 1970 8182.
## 4 Albania 1980 10796.
## 5 Albania 1990 9627.
## 6 Albania 2000 18765.
## 7 Austria 1950 52056.
## 8 Austria 1960 85666.
## 9 Austria 1970 137585.
## 10 Austria 1980 171559.
## # … with 170 more rowsThose two pipes operators are not equivalent so we recommend to read this R-bloggers’ blogpost on pipe’s operators.
References
Wickham, Hadley, and Jennifer Bryan. 2022. R Packages. 2nd ed. https://r-pkgs.org/.